Machine Learning Algorithms are increasingly interesting for analyzing spatial data, especially to derive spatial predictions / for spatial interpolation and to detect spatial patterns. Spatial auto-correlation, especially if still existent in the cross-validation residuals, indicates that the predictions are maybe biased, and this is suboptimal, hence Machine Learning algorithms need to be adjusted to spatial data problems.
COURSE FEATURES
Video lessons
10 case studies
Spatial prediction
LESSONS
Machine-learning based modelling of spatial and spatio-temporal data (introduction)
Remote sensing is a key method in bridging the gap between local observations and spatially comprehensive estimates of environmental variables. For such spatial or spatio-temporal predictions, machine learning algorithms have shown to be a promising tool to identify nonlinear patterns between locally measured and remotely sensed variables. While easy access to user-friendly machine learning libraries fosters their use in environmental sciences, the application of these methods is far from trivial. This holds especially true for spatio-temporal since its dependencies in space and time bear the risk of overfitting and considerable misinterpretation of the model performance. In this introductory lecture I will introduce the idea of using machine-learning for the (remote sensing based) monitoring of the environment and how they can be applied in R via the caret package. In this context error assessment is a crucial topic and I will show the importance of “target-oriented” spatial cross-validation strategies when working with spatio-temporal data to avoid an overoptimistic view on model performances. As spatio-temporal machine-learning models are highly prone to overfitting caused by misleading predictor variables, I will introduce a forward feature selection method that works in conjunction with target-oriented cross-validation from the CAST package. In summary this talk aims at showing how “basic” spatial machine-learning tasks can be performed in R, but also what needs to be considered for more complex spatio-temporal prediction tasks in order to produce scientifically valuable results. Based on this talk, we will go into a practical session on Tuesday, where machine-learning algorithms will be applied to two different spatial and spatio-temporal prediction tasks.
Slides: https://github.com/HannaMeyer/Geostat2018/tree/master/slides
Machine-learning based modelling of spatial and spatio-temporal data
This practical session will base on the introductory lecture on machine-learning based modelling of spatial and spatio-temporal data held on Monday. Two examples will be provided to dive into machine learning for spatial and spatio-temporal data in R:
The first example is a classic remote sensing example dealing with land cover classification at the example of the Banks Peninsula in New Zealand that suffers from spread of the invasive gorse. In this example we will use the random forest classifier via the caret package to learn the relationships between spectral satellite information and provided reference data on the land cover classes. Spatial predictions will then be made to create a map of land use/cover based on the trained model.
As second example, the vignette “Introduction to CAST” is taken from the CAST package. In this example the aim is to model soil moisture in a spatio-temporal way for the cookfarm (http://gsif.r-forge.r-project.org/cookfarm.html). In this example we focus on the differences between different cross-validation strategies for error assessment of spatio-temporal prediction models as well as on the need of a careful selection of predictor variables to avoid overfitting.
- Slides: https://github.com/HannaMeyer/Geostat2018/tree/master/slides
- Exercise A: https://github.com/HannaMeyer/Geostat2018/tree/master/practice/LUCmodelling.html
- Exercise B: https://github.com/HannaMeyer/Geostat2018/tree/master/practice/CAST-intro.html
- Data for Exercise A: https://github.com/HannaMeyer/Geostat2018/tree/master/practice/data/
Required Packages: caret, randomForest, CAST, raster
Suggested further packages: mapview
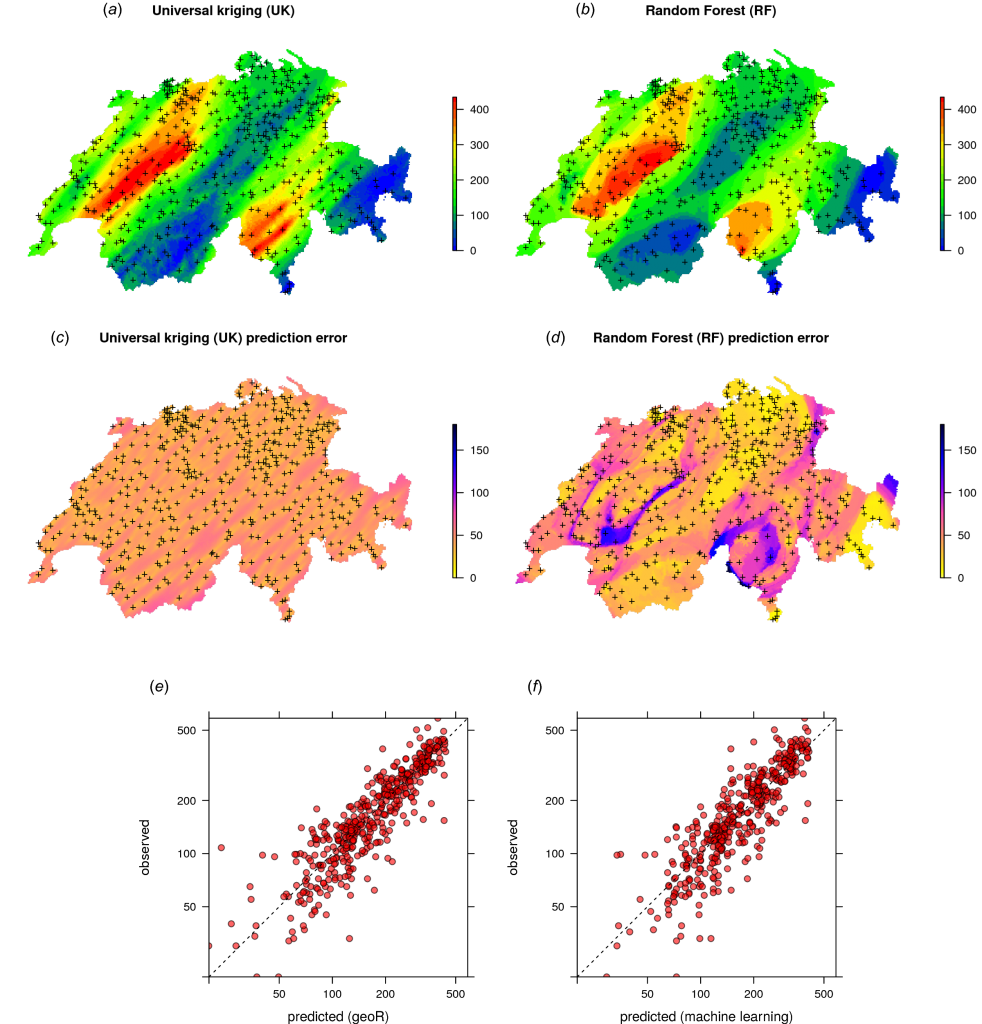
Machine Learning as a generic framework for spatial prediction
Summary: This tutorial explains how to use Random Forest to generate spatial and spatiotemporal predictions (i.e. to make maps from point observations using Random Forest). Spatial auto-correlation, especially if still existent in the cross-validation residuals, indicates that the predictions are maybe biased, and this is sub-optimal. To account for this, we use Random Forest (as implemented in the ranger package) in combination with geographical distances to sampling locations to fit models and predict values.
Tutorials: RFsp — Random Forest for spatial data (https://github.com/thengl/GeoMLA)
Reference:
Hengl T, Nussbaum M, Wright MN, Heuvelink GBM, Gräler B. (2018) Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 6:e5518 https://doi.org/10.7717/peerj.5518
Computing with large rasters in R: tiling, parallelization, optimization
Processing large spatial data in a programming environment such as R is not trivial. Even if you use powerful computing infrastructure, it might take careful programming to be able to process large spatial data. The reasons why R has not been recommended as a programming environment for large data were: (a) R does not handle well large objects in the working memory, and (b) for many existing functions parallelization is not implemented automatically but has to be added through additional programming. This tutorial demonstrates how to use R to read, process and create large spatial (raster) data sets. To obtain the data sets used in the tutorial and R code please refer to the github repository.
Tutorials: Mapping Potential Natural Vegetation (https://github.com/Envirometrix/BigSpatialDataR)
Documentation (slides): https://github.com/Envirometrix/BigSpatialDataR/blob/master/tex/Processing_large_rasters_R.pdf
The importance of spatial cross-validation in predictive modeling
In the first part of this tutorial we will demonstrate the importance of spatial cross-validation and introduce mlr building blocks. We will assess the predictive performance of a random forest model that predicts the floristic composition of Mt. Mongón in northern Peru. In the second part of the tutorial, we will show one way of assessing optimal hyperparameters which are subsequently used for the predictive mapping of the floristic composition.
Software requirements: RStudio, R and its packages mlr, parallelMap, sf, raster, dplyr, mapview, vegan
Materials: The code and presentations can be found in the spatial_cv folders in our GeoStats geocompr repository (https://github.com/geocompr/geostats_18).
References:
- Lovelace, R., Nowosad, J., and J. Muenchow (forthcoming). Geocomputation with R. Chapter 11: Statistical Learning for geographic data. https://bookdown.org/robinlovelace/geocompr/spatial-cv.html. CRS Press.
- Lovelace, R., Nowosad, J., and J. Muenchow (forthcoming). Geocomputation with R. Chapter 14: Ecological use case. https://bookdown.org/robinlovelace/geocompr/eco.html. CRS Press.
- Probst, P., Wright, M., & Boulesteix, A.-L. (2018). Hyperparameters and Tuning Strategies for Random Forest. ArXiv:1804.03515 [Cs, Stat]. Retrieved from http://arxiv.org/abs/1804.03515
- Schratz, P., Muenchow, J., Iturritxa, E., Richter, J., & Brenning, A. (2018). Performance evaluation and hyperparameter tuning of statistical and machine-learning models using spatial data. ArXiv:1803.11266 [Cs, Stat]. Retrieved from http://arxiv.org/abs/1803.11266
Tutorials: Mapping Potential Natural Vegetation (https://github.com/Envirometrix/BigSpatialDataR)
Documentation (slides): https://github.com/Envirometrix/BigSpatialDataR/blob/master/tex/Processing_large_rasters_R.pdf